-
Government
City Services-
DepartmentsCity Manager Administration City Clerk Finance Fire Department Human Resources Library Parks Culture and Recreation Planning Ports and Harbors Public Safety Public Utilities Public Works
-
Mayor & City CouncilMayor & Council Agendas and Minutes Meeting Schedule Public Comment at Meetings Ordinances Resolutions Trip Reports City Code Annual Budget Lobbyist Reports Staff Directory
-
Committees & CommissionsHistoric Preservation Commission Library Advisory Committee Parks, Culture & Recreation Committee Planning Commission Apply to ServeElection InformationRegistering to Vote Polling Location Absentee Voting FAQ

Did You Know?
Did you know that the waters around Unalaska Island are one of the few places in the world to find the rare Whiskered Auklet?
-
-
Commerce
Port/Business-
PortPort Department Dock Facilities and Services AirportFisheriesCommercial Fishing Seasons Sport Fishing Seasons Fisheries Links Fisheries Reports Fish Species common in Alaska
-
BusinessBids and RFPs Business Directory Business Licensing Community Profile Economy Forms and Permits Shipping & Cargo Taxes Unalaska Visitors Bureau
-
TaxesPersonal Property Taxes Real Property Taxes Sales Taxes

Did You Know?
Did you know that Unalaska/Dutch Harbor was bombed by the Japanese during WWII?
-
-
Community
Residents/Visitors-
About UnalaskaCommunity Profile Economy Frequently Asked Questions History Is it Unalaska or Dutch Harbor? Passenger Transportation Photo Galleries Plants Population Shipping and Cargo Weather Wildlife WWII in the Aleutians
-
VisitorsVisitor Bureau Library Museum of the Aleutians Parks & Trails Passenger Transportation Sport Fishing Seasons Things To See & Do Tour Operators WWII National Historic Area and Visitor Center
-
ResidentsAnnual Festivals and Events Community Calendar Emergency Preparedness Health Care Library Non Profits & Community Services Parks & Trails Recreation Schools Sport Fishing Seasons
Did You Know?
Did you know that Unalaska is 800 miles from Anchorage and farther West than Hawaii?
-
-
I want to...
Help Center-
ApplyJob Board/Commission Building Permit Burn Permit Business License Fireworks Permit Pet LicenseContactStaff DirectoryRequestPublic Records Public Safety Records Utility Service
-
PaymentsUtilities TaxesRegisterVoter Registration Vehicle RegistrationReportBuilding Code Violation Road Maintenance Issue Street Light Outage Animal Control Issues
-
ViewAgendas and Minutes Annual Budget Bids & RFPs Code of Ordinances Calendar of Meetings Community Events Documents, Reports, & Presentations Forms, Permits & Applications Property Parcel Info Schedule of Fees & Charges

Did You Know?
That Unalaska’s Port of Dutch Harbor is the #1 Commercial Fishing Port in the United States, based on quantity of the catch and has been for more than 25 years?
Find Out More-
SARS-CoV-2 Wastewater Test Protocols
SARS-CoV-2 Wastewater Test Protocols
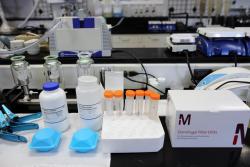
Viral precipitation, RNA extraction, reverse transcription and quantitative PCR (qPCR)
24-hour composite samples of raw sewage were collected during high flow times at two lift stations and the City of Unalaska Waste Water Treatment Plant Influent and stored at 4C. On average 12 grab samples were collected over a 24-hour period.
Samples were mixed well and added to 50 mL centrifuge tubes. Tubes were centrifuged at 8300G for 30 minutes with no breaking force. Supernatant (40 mL) was carefully decanted and mixed with 4g of Polyethylene glycol 8000 and 0.9 g NaCL and centrifuged at 13,000 G for 2.5 hours to precipitate viral particles. In most samples the 6-tube capacity was repeated 4-5 times (total volume- 960- 1,200 mL). The entire volume of viral pellet was transferred into a single tube and allowed to settle overnight at 4C. Before extraction the settled pellet was spun at 8300G for 30 minutes.
Viral precipitation, RNA extraction, reverse transcription and quantitative PCR (qPCR)
24-hour composite samples of raw sewage were collected during high flow times at two lift stations and the City of Unalaska Waste Water Treatment Plant Influent and stored at 4C. On average 12 grab samples were collected over a 24-hour period.
Samples were mixed well and added to 50 mL centrifuge tubes. Tubes were centrifuged at 8300G for 30 minutes with no breaking force. Supernatant (40 mL) was carefully decanted and mixed with 4g of Polyethylene glycol 8000 and 0.9 g NaCL and centrifuged at 13,000 G for 2.5 hours to precipitate viral particles. In most samples the 6-tube capacity was repeated 4-5 times (total volume- 960- 1,200 mL). The entire volume of viral pellet was transferred into a single tube and allowed to settle overnight at 4C. Before extraction the settled pellet was spun at 8300G for 30 minutes.
Viral Precipitation
Method I
The viral pellet was then resuspended in 1.5 mL Trizol reagent for RNA extraction. cDNA was synthesized by reverse transcription:
10 µL of RNA was mixed with random hexamers and then incubated at 70C for 5 min and 4C for 3 min. After that, the RNA-hexamer mixture was mixed with 5X ProtoScript II buffer (5 µL), 0.1M DTT (2.5 µL), ProtoScript II Reverse Transcriptase (200 U/µl. 1.25 µL), 10 mM dNTP (1.25 ul), RNase Inhibitor (40 U/µl, 0.5ul), and Nuclease-free water (2.5 µl) to a total volume of 25 µl. The mixture was then incubated at 42C for 1 hour and inactivated at 65C for 20 minutes.
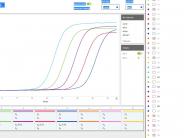
The quantitative PCR (qPCR) was performed with TaqMan assay techniques. The Taqman Fast Advanced Master Mix (4444557, Thermofisher Scientific) was mixed with the primers and water and 2 ul of cDNA template from RT was added. The qPCR reaction was carried out for 48 cycles using Chai Open PCR QPCR Detection System based on the following program: polymerase activation (95C for 2 min), PCR (48 cycles, denature at 95C for 1 s), and anneal/extend at (55C for 30 s).
Method II
The viral pellet was then added to the PowerBead Bead Tube and RNA extracted with the Qiagen RNeasy PowerMicrobiome Kit (Cat. #26000-50). The eluted RNA (10µL-20µL) was immediately used for one-step RT-PCR with Taqman Fast Virus 1-Step Master Mix (Thermofisher, Cat. #4444436) based on the following protocol: Reverse transcription (50 C 10 min), RT inactivation and initial denaturation (95C 20 s), and 48 cycles of denature (95C 1 s) and anneal/extend (55C 30 s).
The primers (N1 and N2) and DNA standards of SARS-CoV-2 nucleocapsid gene were used to quantify the titers of SARS-CoV-2 (Integrated DNA Technologies (IDT) 2019-nCoV CDC EUA qPCR Probe Assay primer/probe mix and 2019-nCoV_N Positive Control plasmid). Two replicates were performed. Positive and negative controls were included in each run as well as internal controls.
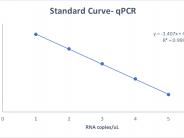
Standard Curve obtained with synthetic SARS-CoV-2 ((Integrated DNA Technologies (IDT)).
MilliQ water was spiked with standard solution of synthetic SARS-CoV-2 ((Integrated DNA Technologies (IDT)) to perform a standard curve. The trend line has a slope of -3.407 and a R value of 0.999. The linear trend between Ct values and RNA copy numbers allowed for quantitative analysis of virus RNA concentration and to track SARS-CoV-2 in the wastewater over time.
QA/QC
The extracted RNA had an ideal result of high concentration and a purity of OD260/OD280: 2.0
Internal Controls
The method for recovery and concentration of SARS-CoV-2 from wastewater was validated and standardized using an internal process control and an internal amplification control. Pepper Mild Mottle Virus (PPMoV), the most abundant RNA virus in human feces, was used as an internal standardization control (Promega Wastewater SARS-CoV-2 N1 qPCR kit).
Additionally, an extraction control containing synthetic SARS-CoV-2 from the Seracare AccuPlex SARS-CoV-2 Verification Panel (0505-0168) was used.
Methods developed for testing SARS-CoV-2 will be useful for testing other pathogens or another future pandemic.
Click any thumbnail image to view a slideshow